
New paper about the Neurodata Without Borders (NWB) standards effort
7 October 2022The Neurodata Without Borders (NWB) team recently published a paper in eLife, “The Neurodata Without Borders ecosystem for neurophysiological data science” that describes their 8 year long effort to develop a data standard and a surrounding software ecosystem for neurophysiology data.
The NWB effort began in mid-2014, when INCF, Kavli Foundation, GE, Janelia Farm, and Allen Brain Institute initiated Neurodata Without Borders as a consortium of researchers and foundations with a shared interest in breaking down the obstacles to data sharing in neuroscience. Their first pilot project was a standardized file format for neurophysiology data, Neurodata Without Borders: Neurophysiology (NWB:N).
Now commonly referred to as just “NWB”, the data standard is based on the HDF5 format, using recommendations developed by an INCF working group under the previous INCF Program on Standards for Data Sharing. The format contains all of the measurements for a single neurophysiology experiment, and all metadata needed to understand the data. NWB was endorsed as an INCF standard in 2020.
The issue that NWB tries to address is that neurophysiology datasets are both very big and complex, making them tricky to share or reuse. Neurophysiology experiments often contain multiple simultaneous streams of data from neural activity, sensory stimuli, behavioral tracking, and direct neural modulation. Often multiple neurophysiology recording modalities are used at the same time (e.g. electrophysiology and optophysiology). The raw data input requires processing, which expands the multiplicity of data types to describe and store even more. As a consequence, with all neurophysiology labs describing and storing their datasets differently, cross-lab collaborations and data reuse was nearly impossible.
Lack of user-friendly tools is a well-known bottleneck to standards adoption in the wider community. The NWB ecosystem has several other components that make the standard easier to implement for tool developers.
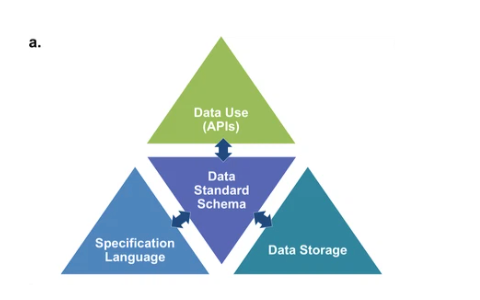
Figure 3a from the paper showing the main components of the NWB software stack: 1) the specification language to describe data standards 2) the data standard schema, which uses the specification language to formally define the data standard; 3) the data storage for translating the data primitives (e.g., groups & datasets) described by the schema to/from disk 4) the APIs that enable users to easily read and write data using the standard.
Two API toolkits have been developed, for the two most common programming languages in neurophysiology: Matlab and Python. The MatNWB toolkit primarily targets data conversion and analysis. The PyNWB toolkit also targets integration with data archives and web technologies, and is used heavily for development of extensions and exploration of new technologies, such as alternate storage backends and parallel computing libraries.
A neat bonus is that the interoperability afforded by the PyNWB builders also allows for other storage backends. There is already a prototype for storing NWB in the Zarr format (a community-developed next-generation file format that supports storage in the cloud and has efficient I/O for parallel computing applications).
Resources:
- Paper: “The Neurodata Without Borders ecosystem for neurophysiological data science”
- Learn more about NWB in the NWB collection on TrainingSpace
- Find NWB datasets in the DANDI archive
- Find NWB questions, answers and discussions on Neurostars

